publications
I work at the intersection of AI and the life sciences, especially AI for Proteins and structure‑based drug discovery. Below are my publications by area. Earlier work in mobile computing is also included.
AI for Science
AI for Protein
2025
-
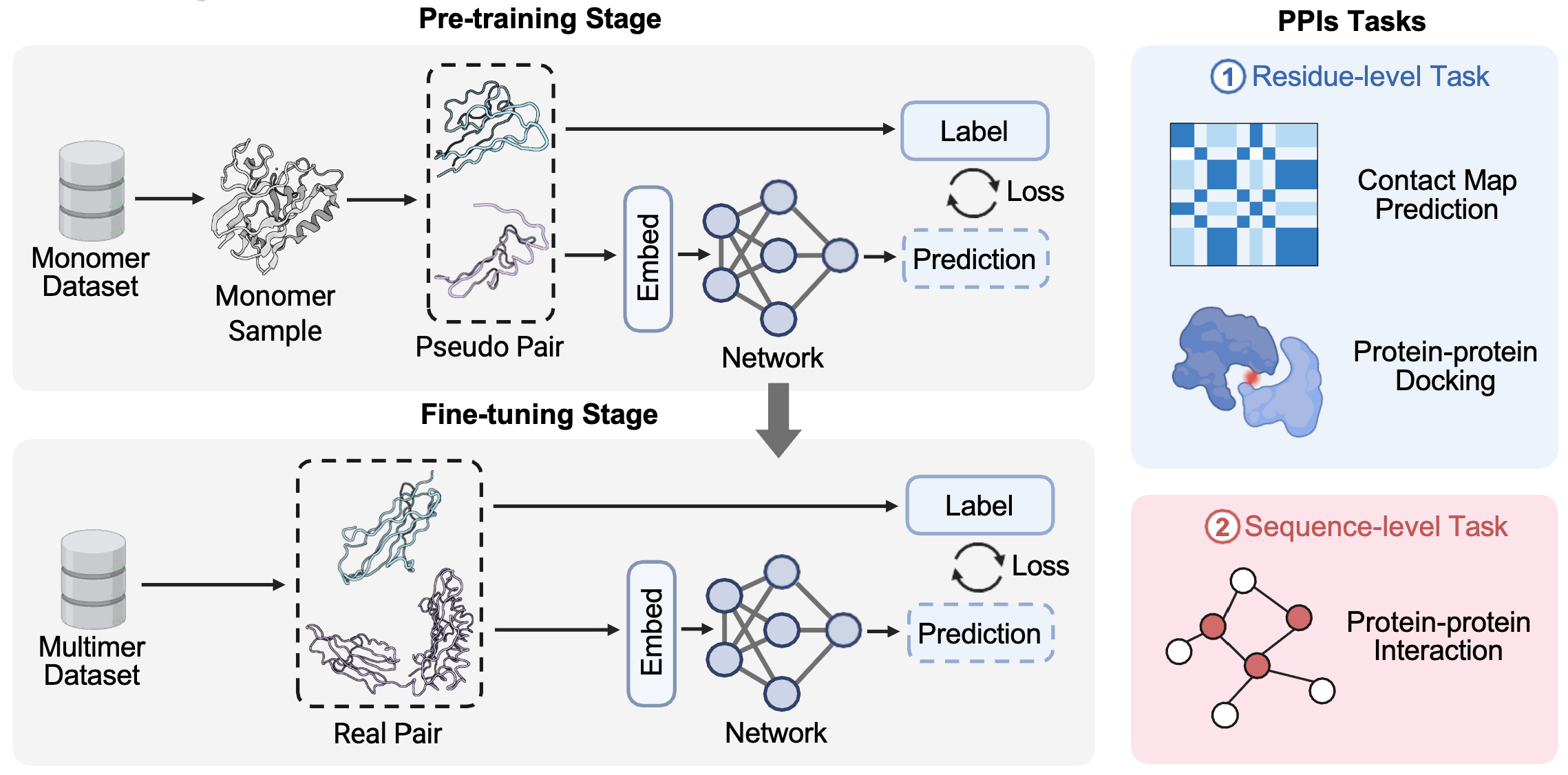 Improving protein and protein interactions using pseudo-dimers derived from monomeric proteinsHao Du*, Xinzhe Zheng*, Yuchen Ren*, and 5 more authorsIn under review, 2025
Improving protein and protein interactions using pseudo-dimers derived from monomeric proteinsHao Du*, Xinzhe Zheng*, Yuchen Ren*, and 5 more authorsIn under review, 2025 -
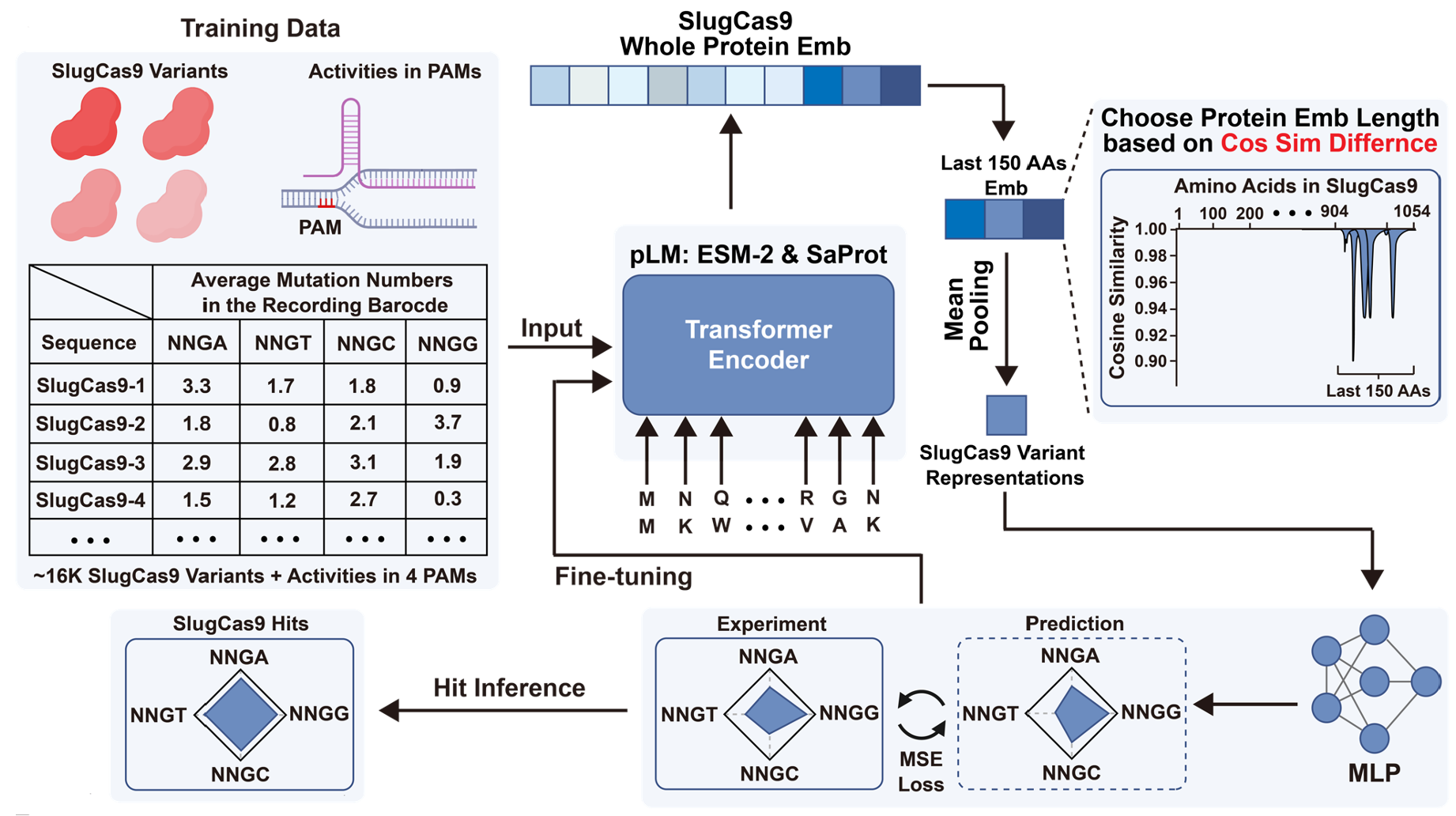 Sequence Display: Generating Large-Scale Sequence–Activity Datasets to Advance Universal Protein EvolutionLinqi Cheng*, Xinzhe Zheng*, Shiyu (Jason) Jiang*, and 12 more authorsIn under review, 2025
Sequence Display: Generating Large-Scale Sequence–Activity Datasets to Advance Universal Protein EvolutionLinqi Cheng*, Xinzhe Zheng*, Shiyu (Jason) Jiang*, and 12 more authorsIn under review, 2025 -
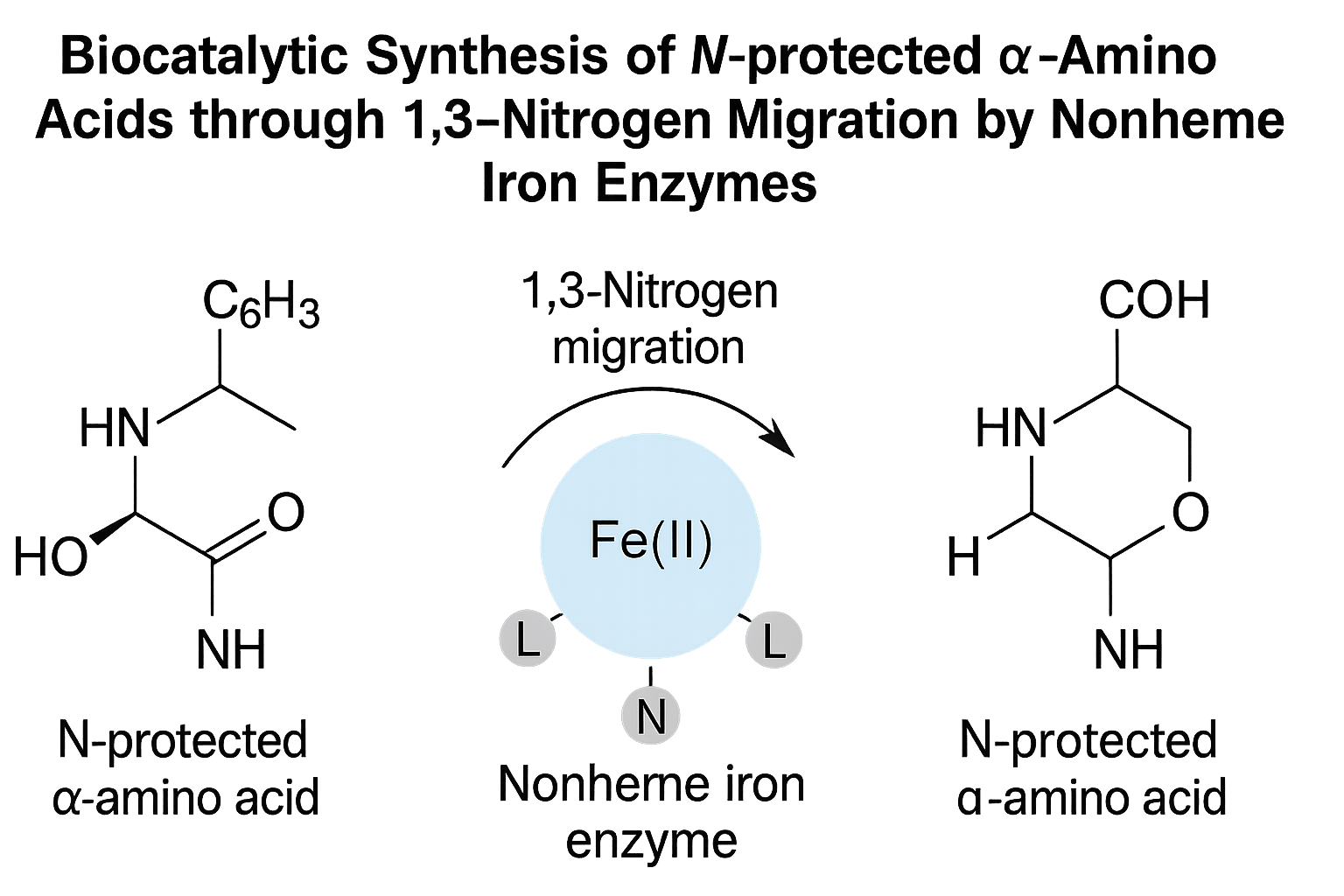 Biocatalytic Synthesis of N-protected α-Amino Acids through 1,3-Nitrogen Migration by Nonheme Iron EnzymesTeng Yuan*, Mengxi Zhang*, Linqi Cheng, and 4 more authorsIn under review, 2025
Biocatalytic Synthesis of N-protected α-Amino Acids through 1,3-Nitrogen Migration by Nonheme Iron EnzymesTeng Yuan*, Mengxi Zhang*, Linqi Cheng, and 4 more authorsIn under review, 2025 -
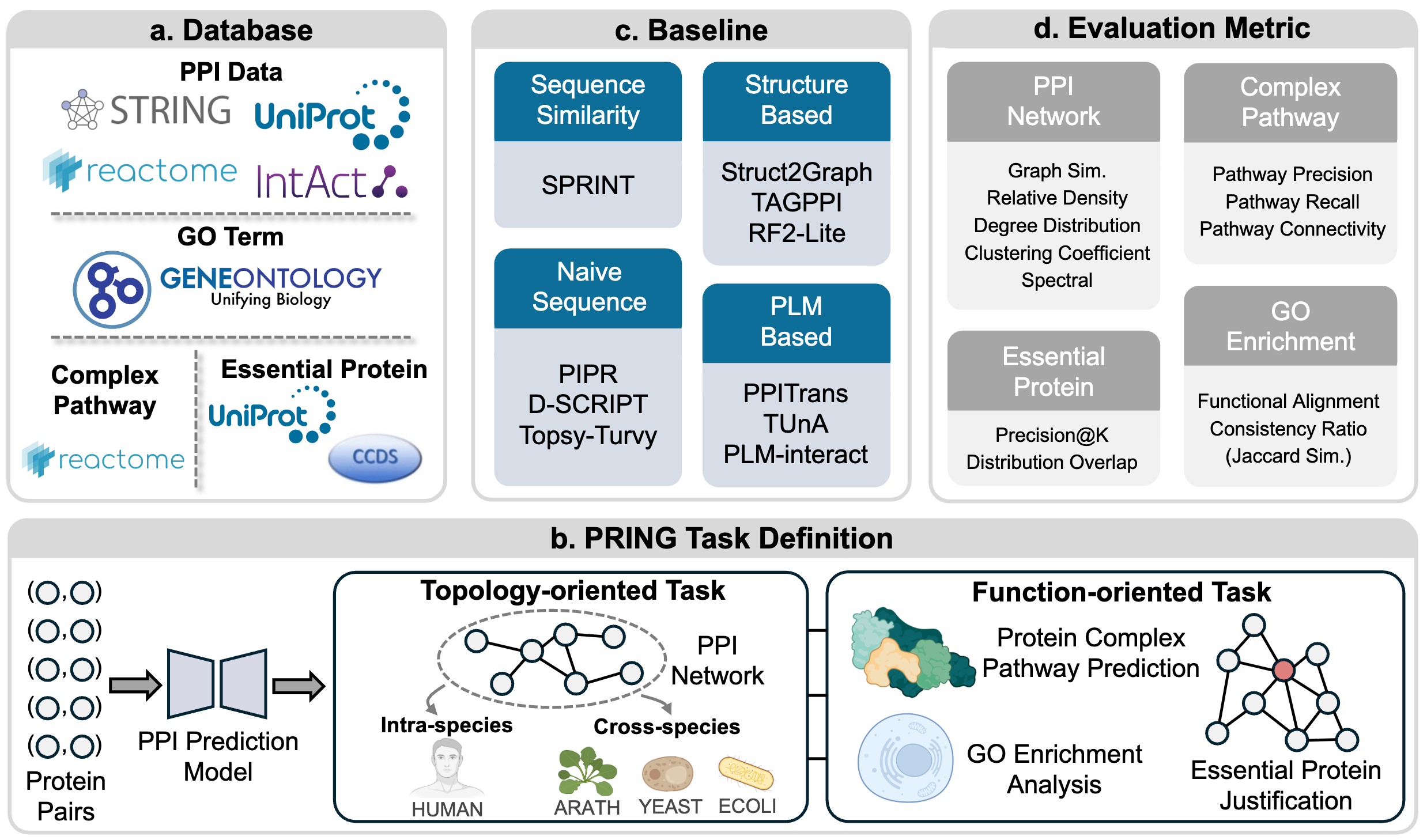 PRING: Rethinking Protein-Protein Interaction Prediction from Pairs to GraphsXinzhe Zheng*, Hao Du*, Fanding Xu*, and 9 more authorsIn NeurIPS Datasets and Benchmarks Track, 2025
PRING: Rethinking Protein-Protein Interaction Prediction from Pairs to GraphsXinzhe Zheng*, Hao Du*, Fanding Xu*, and 9 more authorsIn NeurIPS Datasets and Benchmarks Track, 2025Deep learning-based computational methods have achieved promising results in predicting protein-protein interactions (PPIs). However, existing benchmarks predominantly focus on isolated pairwise evaluations, overlooking a model’s capability to reconstruct biologically meaningful PPI networks, which is crucial for biology research. To address this gap, we introduce PRING, the first comprehensive benchmark that evaluates PRotein-protein INteraction prediction from a Graph-level perspective. PRING curates a high-quality, multi-species PPI network dataset comprising 21,484 proteins and 186,818 interactions, with well-designed strategies to address both data redundancy and leakage. Building on this golden-standard dataset, we establish two complementary evaluation paradigms: (1) topology-oriented tasks, which assess intra and cross-species PPI network construction, and (2) function-oriented tasks, including protein complex pathway prediction, GO module analysis, and essential protein justification. These evaluations not only reflect the model’s capability to understand the network topology but also facilitate protein function annotation, biological module detection, and even disease mechanism analysis. Extensive experiments on four representative model categories, consisting of sequence similarity-based, naive sequence-based, protein language model-based, and structure-based approaches, demonstrate that current PPI models have potential limitations in recovering both structural and functional properties of PPI networks, highlighting the gap in supporting real-world biological applications. We believe PRING provides a reliable platform to guide the development of more effective PPI prediction models for the community. The dataset and source code of PRING are available at https://github.com/SophieSarceau/PRING.
@inproceedings{zheng2025pring, title = {{PRING}: Rethinking Protein-Protein Interaction Prediction from Pairs to Graphs}, author = {Zheng, Xinzhe and Du, Hao and Xu, Fanding and Li, Jinzhe and Liu, Zhiyuan and WenkangWang and Chen, Tao and Ouyang, Wanli and Li, Stan Z. and Lu, Yan and Dong, Nanqing and Zhang, Yang}, booktitle = {NeurIPS Datasets and Benchmarks Track}, year = {2025}, url = {https://openreview.net/forum?id=mHCOVlFXTw}, keywords = {ai4protein}, } -
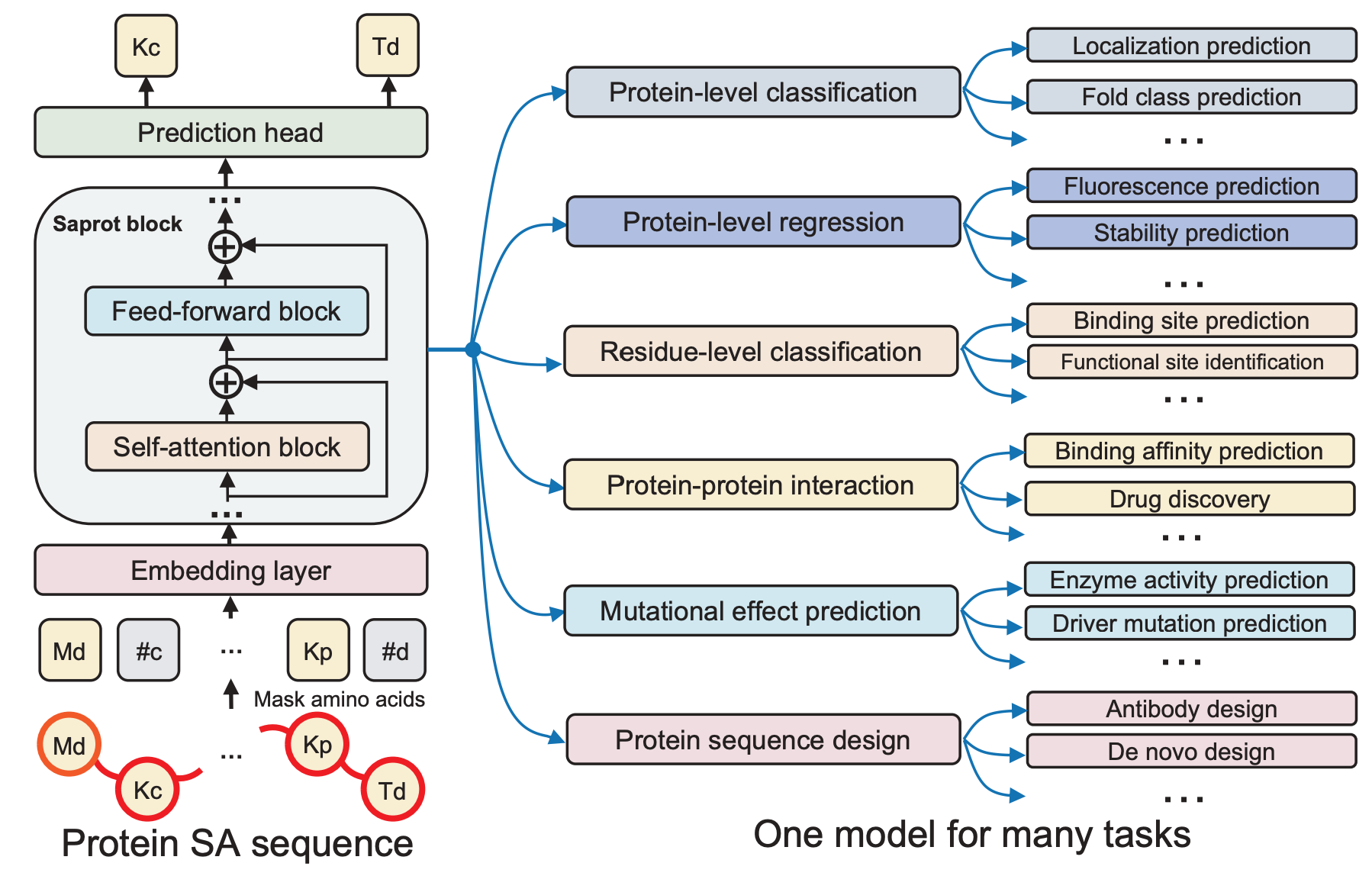 Democratizing protein language model training, sharing and collaborationJin Su, Zhikai Li, Chenchen Han, and 11 more authorsNature Biotechnology Brief Communications, 2025
Democratizing protein language model training, sharing and collaborationJin Su, Zhikai Li, Chenchen Han, and 11 more authorsNature Biotechnology Brief Communications, 2025Training and deploying large-scale protein language models typically requires deep machine learning expertise—a barrier for researchers outside this field. SaprotHub overcomes this challenge by offering an intuitive platform that facilitates training and prediction as well as storage and sharing of models. Here we provide the ColabSaprot framework built on Google Colab, which potentially powers hundreds of protein training and prediction applications, enabling researchers to collaboratively build and share customized models.
AI for Drug Discovery
2026
-
 Apo2Mol: 3D Molecule Generation via Dynamic Pocket-Aware Diffusion ModelsXinzhe Zheng, Shiyu Jiang, Gustovo Seabra, and 2 more authorsIn AAAI, 2026
Apo2Mol: 3D Molecule Generation via Dynamic Pocket-Aware Diffusion ModelsXinzhe Zheng, Shiyu Jiang, Gustovo Seabra, and 2 more authorsIn AAAI, 2026
Other
2025
-
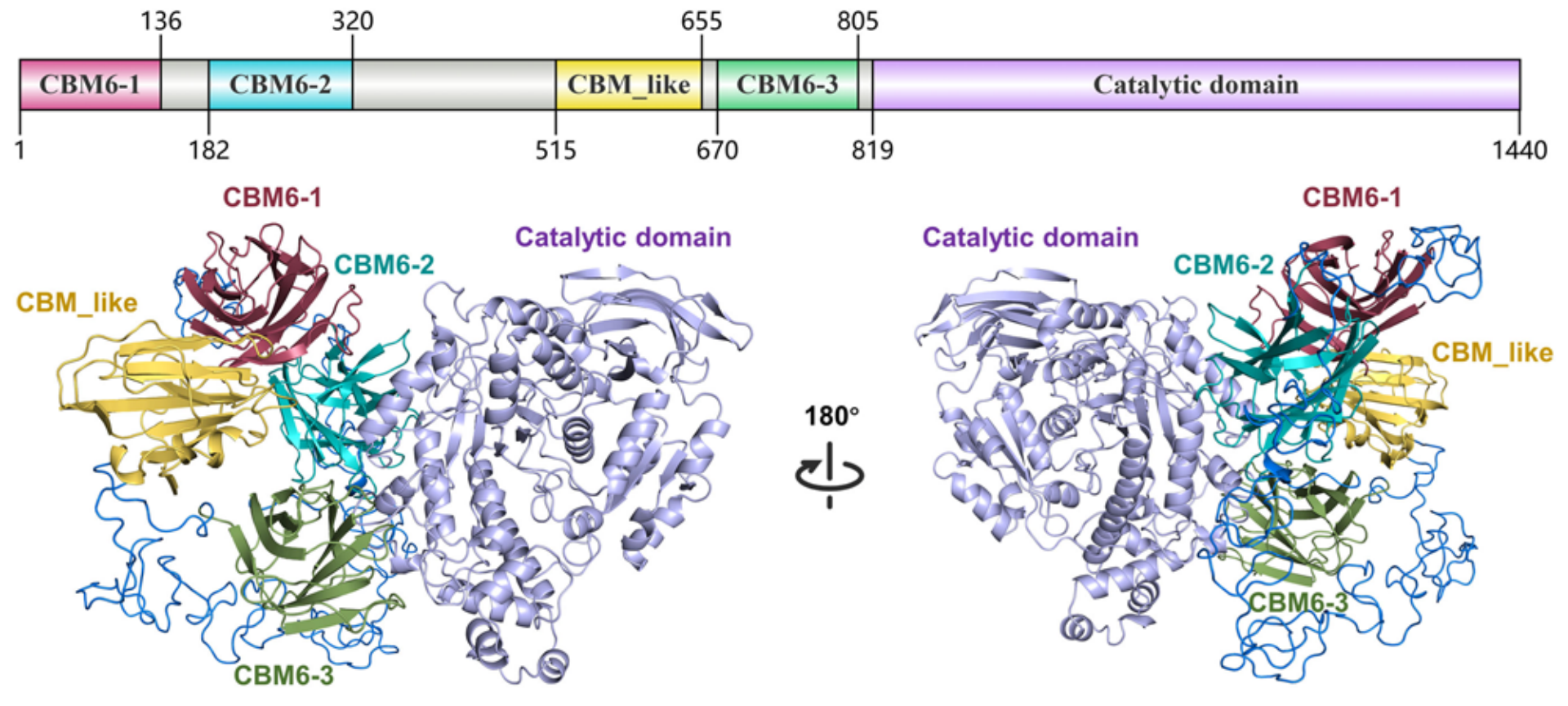 Structure-Informed Insights into Catalytic Mechanism and Multidomain Collaboration in α-Agarase CmAgaYuxian You, Bee Koon Gan, Min Luo, and 8 more authorsJournal of Agricultural and Food Chemistry, 2025
Structure-Informed Insights into Catalytic Mechanism and Multidomain Collaboration in α-Agarase CmAgaYuxian You, Bee Koon Gan, Min Luo, and 8 more authorsJournal of Agricultural and Food Chemistry, 2025α-Agarases are glycoside hydrolases that cleave α-1,3-glycosidic bonds in agarose to produce bioactive agarooligosaccharides. Despite their great industrial potential, the structures and functional mechanisms of α-agarases remain unclear due to their complex and flexible architecture. Here, we investigated the structure-based catalytic mechanism of α-agarase CmAga from Catenovulum maritimum STB14 by integrated Cryo-EM and AlphaFold2. D994 and E1129 were identified as catalytic residues, with E1129 selectively recognizing α-1,3-glycosidic bonds. Y858, W1201, Y1164, and W1166 facilitate preferential substrate binding at the −3 ∼ +3 subsites. Molecular dynamics simulations and neural relational inference modeling revealed a cooperative mechanism involving the catalytic domain (CD) and four carbohydrate-binding modules (CBMs), with CBM6–1 and CBM6–2 capturing substrates, CBM_like transferring them to the CD, and CBM6–3 stabilizing the active site. D149 and L608 served as pivotal nodes within the interdomain communication pathways. These insights provide a foundation for mechanistic investigations and rational engineering of carbohydrate-active enzymes (CAZymes) with multiple CBMs.
@article{you2025structure, title = {Structure-Informed Insights into Catalytic Mechanism and Multidomain Collaboration in $\alpha$-Agarase CmAga}, author = {You, Yuxian and Gan, Bee Koon and Luo, Min and Zheng, Xinzhe and Dong, Nanqing and Tian, Yixiong and Li, Caiming and Kong, Haocun and Gu, Zhengbiao and Yang, Daiwen and Li, Zhaofeng}, journal = {Journal of Agricultural and Food Chemistry}, volume = {73}, number = {13}, pages = {7975--7989}, year = {2025}, publisher = {ACS Publications}, keywords = {ai4s}, } -
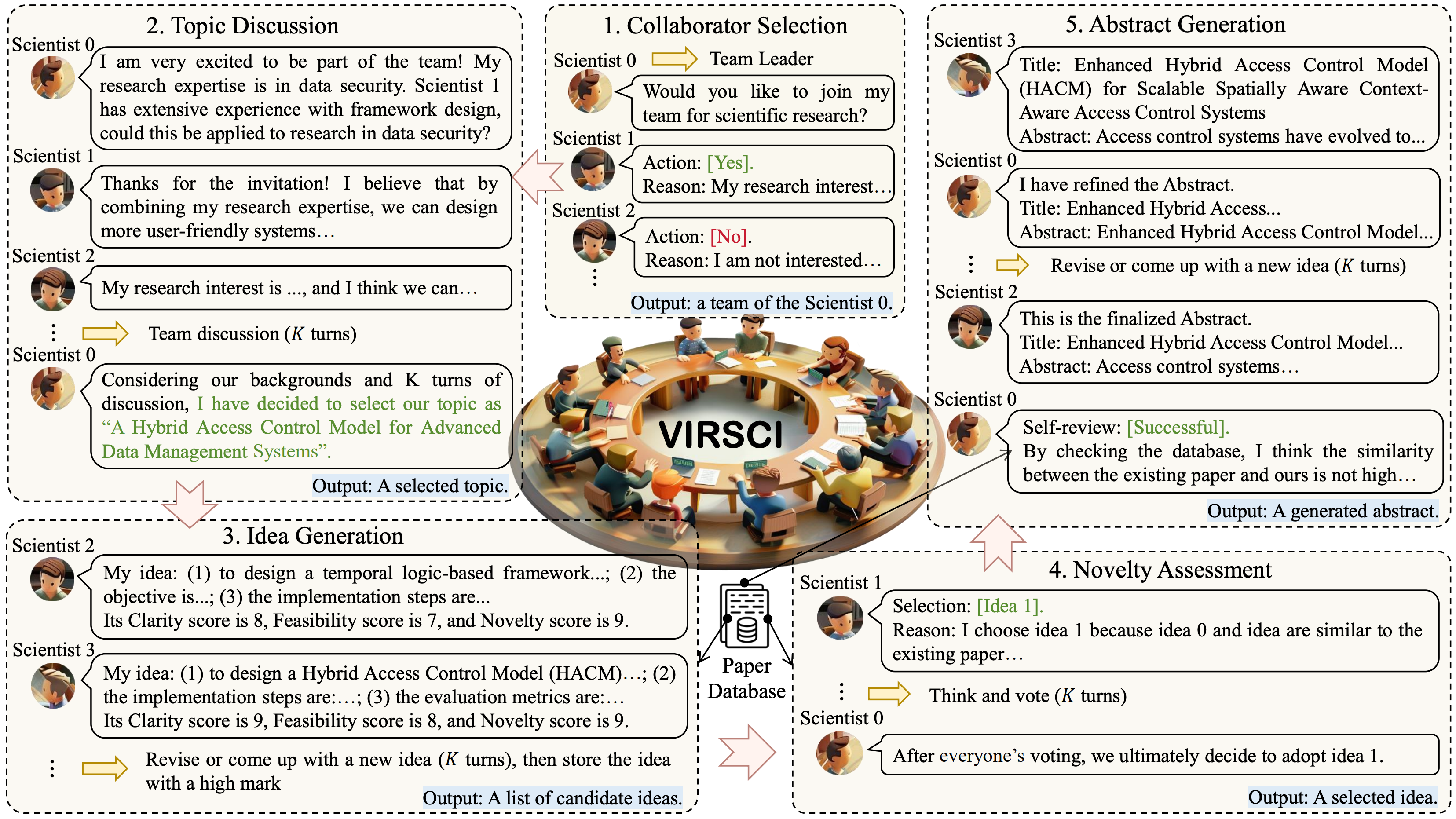 Many Heads Are Better Than One: Improved Scientific Idea Generation by A LLM-Based Multi-Agent SystemHaoyang Su*, Renqi Chen*, Shixiang Tang†, and 10 more authorsIn ACL, 2025
Many Heads Are Better Than One: Improved Scientific Idea Generation by A LLM-Based Multi-Agent SystemHaoyang Su*, Renqi Chen*, Shixiang Tang†, and 10 more authorsIn ACL, 2025The rapid advancement of scientific progress requires innovative tools that can accelerate knowledge discovery. Although recent AI methods, particularly large language models (LLMs), have shown promise in tasks such as hypothesis generation and experimental design, they fall short of replicating the collaborative nature of real-world scientific practices, where diverse experts work together in teams to tackle complex problems. To address the limitations, we propose an LLM-based multi-agent system, i.e., Virtual Scientists (VIRSCI), designed to mimic the teamwork inherent in scientific research. VIRSCI organizes a team of agents to collaboratively generate, evaluate, and refine research ideas. Through comprehensive experiments, we demonstrate that this multi-agent approach outperforms the state-of-the-art method in producing novel scientific ideas. We further investigate the collaboration mechanisms that contribute to its tendency to produce ideas with higher novelty, offering valuable insights to guide future research and illuminating pathways toward building a robust system for autonomous scientific discovery. The code is available at https://github.com/open-sciencelab/Virtual-Scientists.
@inproceedings{su-etal-2025-many, title = {Many Heads Are Better Than One: Improved Scientific Idea Generation by A {LLM}-Based Multi-Agent System}, author = {Su, Haoyang and Chen, Renqi and Tang, Shixiang and Yin, Zhenfei and Zheng, Xinzhe and Li, Jinzhe and Qi, Biqing and Wu, Qi and Li, Hui and Ouyang, Wanli and Torr, Philip and Zhou, Bowen and Dong, Nanqing}, booktitle = {ACL}, year = {2025}, address = {Vienna, Austria}, publisher = {Association for Computational Linguistics}, url = {https://aclanthology.org/2025.acl-long.1368/}, doi = {10.18653/v1/2025.acl-long.1368}, keywords = {ai4s}, }
2024
-
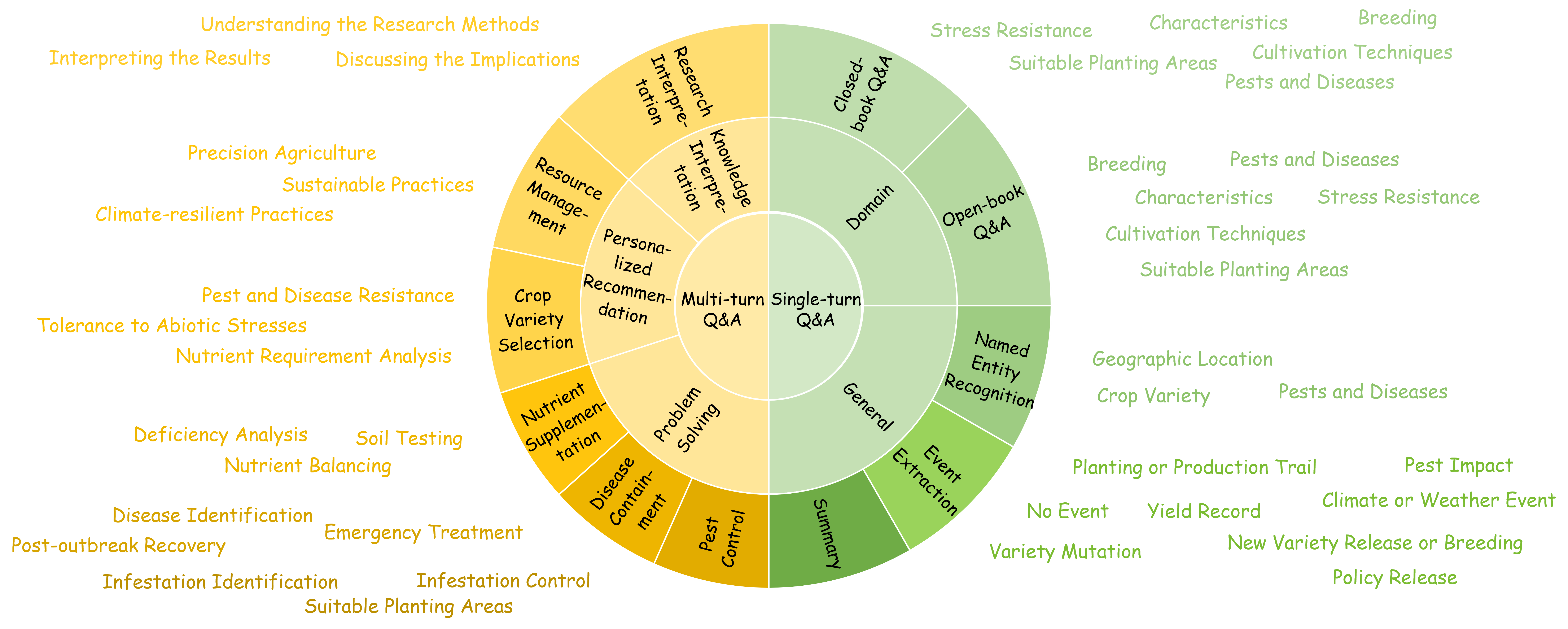 Empowering and Assessing the Utility of Large Language Models in Crop ScienceHang Zhang*, Jiawei Sun*, Renqi Chen*, and 11 more authorsIn NeurIPS Datasets and Benchmarks Track, 2024
Empowering and Assessing the Utility of Large Language Models in Crop ScienceHang Zhang*, Jiawei Sun*, Renqi Chen*, and 11 more authorsIn NeurIPS Datasets and Benchmarks Track, 2024Large language models (LLMs) have demonstrated remarkable efficacy across knowledge-intensive tasks. Nevertheless, their untapped potential in crop science presents an opportunity for advancement. To narrow this gap, we introduce CROP, which includes a novel instruction tuning dataset specifically designed to enhance LLMs’ professional capabilities in the crop science sector, along with a benchmark that serves as a comprehensive evaluation of LLMs’ understanding of the domain knowledge. The CROP dataset is curated through a task-oriented and LLM-human integrated pipeline, comprising 210,038 single-turn and 1,871 multi-turn dialogues related to crop science scenarios. The CROP benchmark includes 5,045 multiple-choice questions covering three difficulty levels. Our experiments based on the CROP benchmark demonstrate notable enhancements in crop science-related tasks when LLMs are fine-tuned with the CROP dataset. To the best of our knowledge, CROP dataset is the first-ever instruction tuning dataset in the crop science domain. We anticipate that CROP will accelerate the adoption of LLMs in the domain of crop science, ultimately contributing to global food production.
@inproceedings{zhang2024empowering, title = {Empowering and Assessing the Utility of Large Language Models in Crop Science}, author = {Zhang, Hang and Sun, Jiawei and Chen, Renqi and Liu, Wei and Yuan, Zhonghang and Zheng, Xinzhe and Wang, Zhefan and Yang, Zhiyuan and Yan, Hang and Zhong, Han-Sen and Wang, Xiqing and Ouyang, Wanli and Yang, Fan and Dong, Nanqing}, booktitle = {NeurIPS Datasets and Benchmarks Track}, year = {2024}, url = {https://openreview.net/forum?id=hMj6jZ6JWU}, keywords = {ai4s}, } -
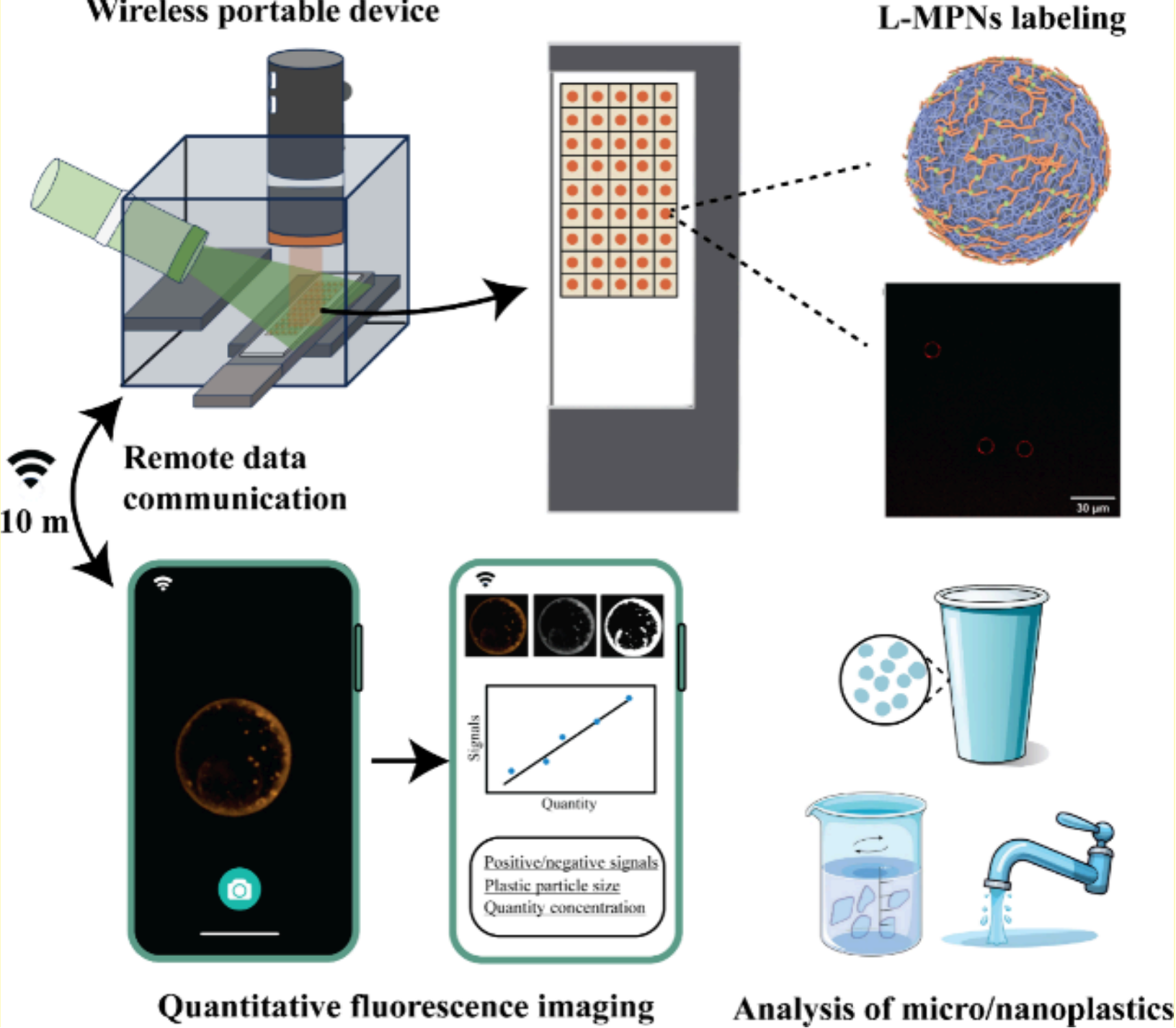 Cost-effective and wireless portable device for rapid and sensitive quantification of micro/nanoplasticsHaoxin Ye, Xinzhe Zheng, Haoming Yang, and 9 more authorsACS sensors, 2024
Cost-effective and wireless portable device for rapid and sensitive quantification of micro/nanoplasticsHaoxin Ye, Xinzhe Zheng, Haoming Yang, and 9 more authorsACS sensors, 2024The accumulation of micro/nanoplastics (MNPs) in ecosystems poses tremendous environmental risks for terrestrial and aquatic organisms. Designing rapid, field-deployable, and sensitive devices for assessing the potential risks of MNPs pollution is critical. However, current techniques for MNPs detection have limited effectiveness. Here, we design a wireless portable device that allows rapid, sensitive, and on-site detection of MNPs, followed by remote data processing via machine learning algorithms for quantitative fluorescence imaging. We utilized a supramolecular labeling strategy, employing luminescent metal–phenolic networks composed of zirconium ions, tannic acid, and rhodamine B, to efficiently label various sizes of MNPs (e.g., 50 nm–10 μm). Results showed that our device can quantify MNPs as low as 330 microplastics and 3.08 × 106 nanoplastics in less than 20 min. We demonstrated the applicability of the device to real-world samples through determination of MNPs released from plastic cups after hot water and flow induction and nanoplastics in tap water. Moreover, the device is user-friendly and operative by untrained personnel to conduct data processing on the APP remotely. The analytical platform integrating quantitative imaging, customized data processing, decision tree model, and low-cost analysis ($0.015 per assay) has great potential for high-throughput screening of MNPs in agrifood and environmental systems.
@article{ye2024cost, title = {Cost-effective and wireless portable device for rapid and sensitive quantification of micro/nanoplastics}, author = {Ye, Haoxin and Zheng, Xinzhe and Yang, Haoming and Kowal, Matthew D. and Seifried, Teresa M. and Singh, Gurvendra Pal and Aayush, Krishna and Gao, Guang and Grant, Edward and Kitts, David and Yada, Rickey Y. and Yang, Tianxi}, journal = {ACS sensors}, volume = {9}, number = {9}, pages = {4662--4670}, year = {2024}, publisher = {ACS Publications}, keywords = {ai4s}, }
AI for Health
2026
-
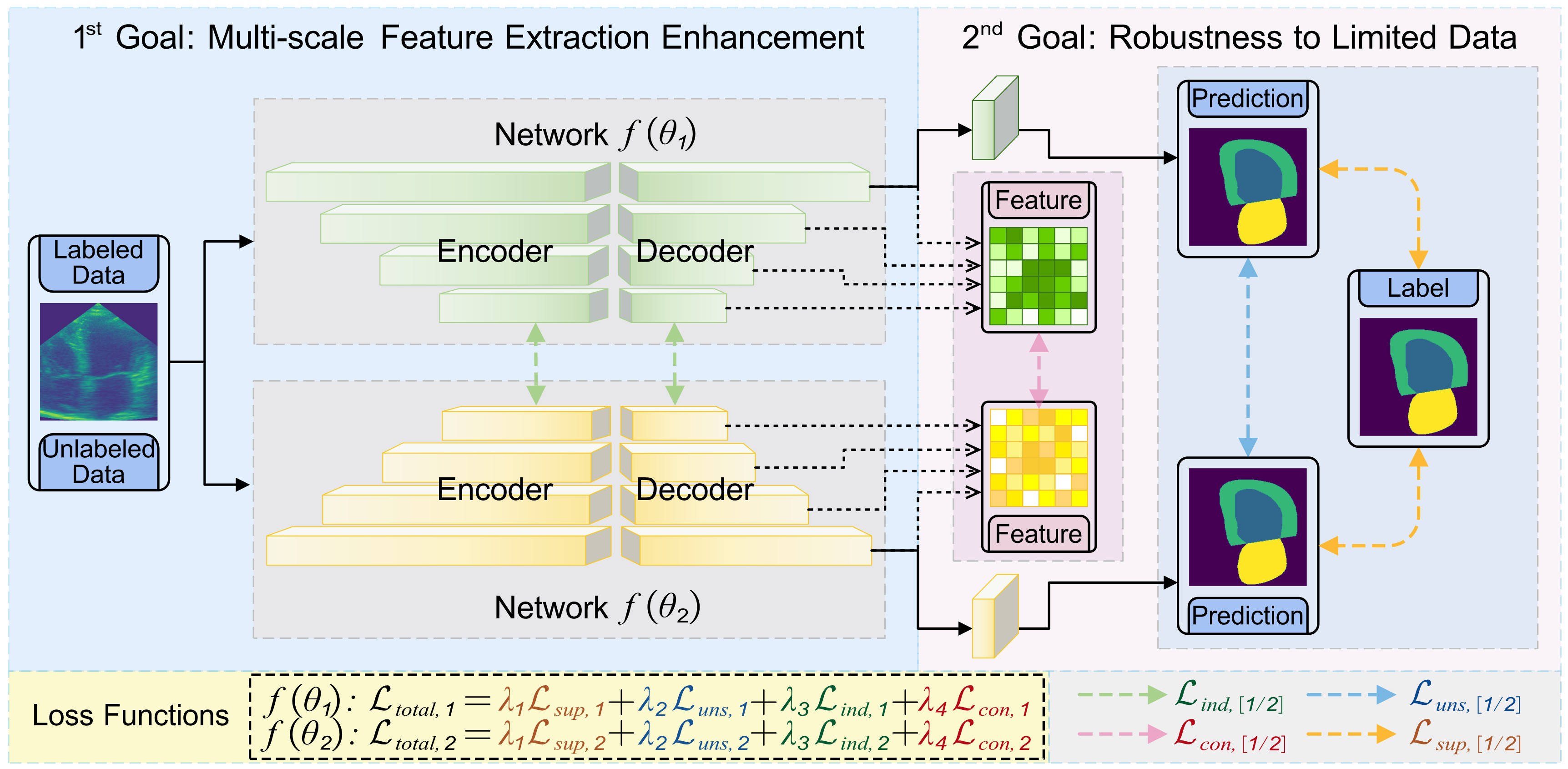 HCS-TNAS: Hybrid Constraint-driven Semi-supervised Transformer-NAS for Ultrasound Image SegmentationRenqi Chen, Xinzhe Zheng, Haoyang Su, and 1 more authorIn AAAI, 2026
HCS-TNAS: Hybrid Constraint-driven Semi-supervised Transformer-NAS for Ultrasound Image SegmentationRenqi Chen, Xinzhe Zheng, Haoyang Su, and 1 more authorIn AAAI, 2026
2025
-
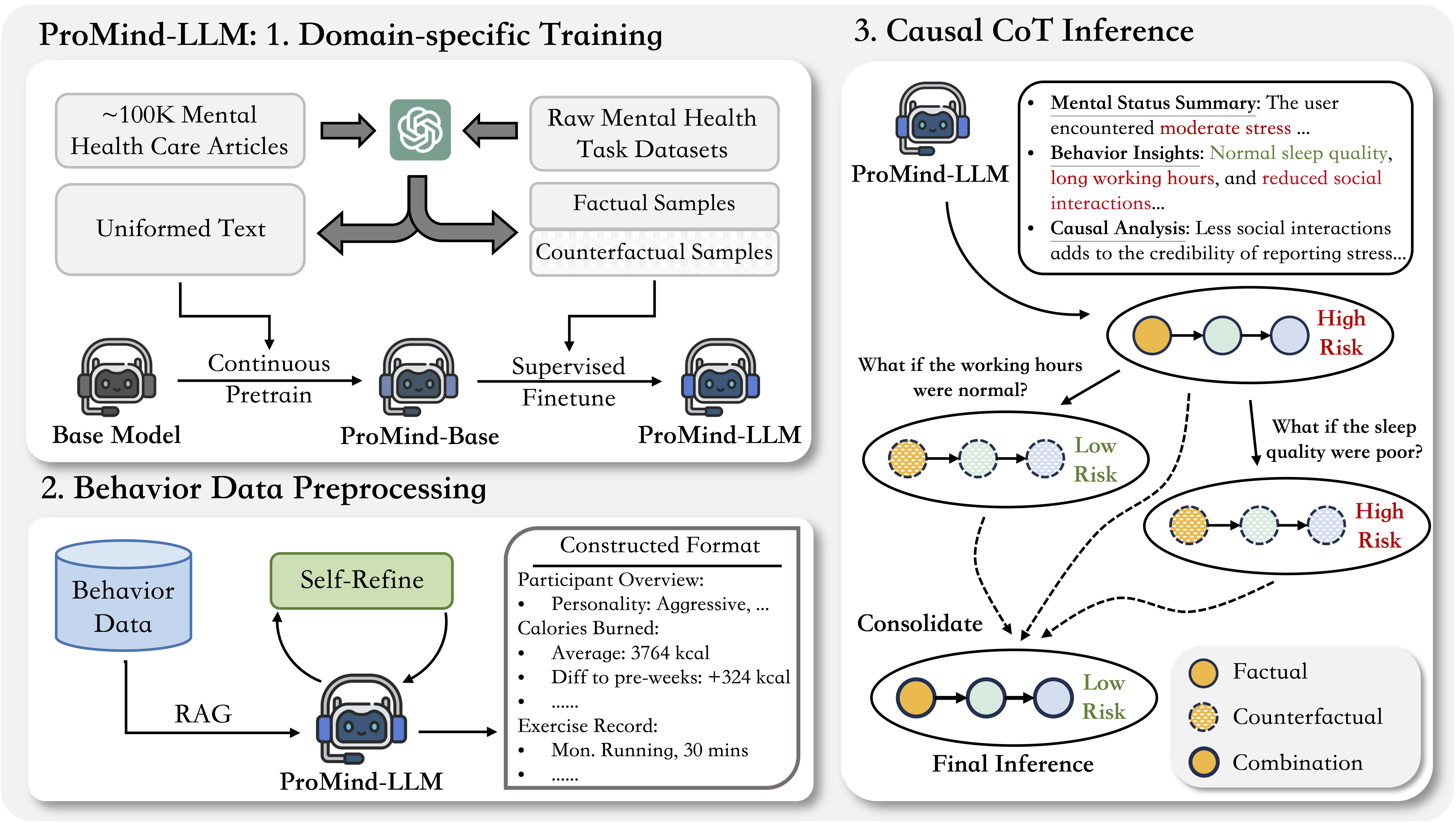 ProMind-LLM: Proactive Mental Health Care via Causal Reasoning with Sensor DataXinzhe Zheng*, Sijie Ji*†, Jiawei Sun*, and 3 more authorsIn ACL Findings, 2025
ProMind-LLM: Proactive Mental Health Care via Causal Reasoning with Sensor DataXinzhe Zheng*, Sijie Ji*†, Jiawei Sun*, and 3 more authorsIn ACL Findings, 2025Mental health risk is a critical global public health challenge, necessitating innovative and reliable assessment methods. With the development of large language models (LLMs), they stand out to be a promising tool for explainable mental health care applications. Nevertheless, existing approaches predominantly rely on subjective textual mental records, which can be distorted by inherent mental uncertainties, leading to inconsistent and unreliable predictions. To address these limitations, this paper introduces ProMind-LLM. We investigate an innovative approach integrating objective behavior data as complementary information alongside subjective mental records for robust mental health risk assessment. Specifically, ProMind-LLM incorporates a comprehensive pipeline that includes domain-specific pretraining to tailor the LLM for mental health contexts, a self-refine mechanism to optimize the processing of numerical behavioral data, and causal chain-of-thought reasoning to enhance the reliability and interpretability of its predictions. Evaluations of two real-world datasets, PMData and Globem, demonstrate the effectiveness of our proposed methods, achieving substantial improvements over general LLMs. We anticipate that ProMind-LLM will pave the way for more dependable, interpretable, and scalable mental health case solutions.
@inproceedings{zheng-etal-2025-promind, title = {{P}ro{M}ind-{LLM}: Proactive Mental Health Care via Causal Reasoning with Sensor Data}, author = {Zheng, Xinzhe and Ji, Sijie and Sun, Jiawei and Chen, Renqi and Gao, Wei and Srivastava, Mani}, booktitle = {ACL Findings}, year = {2025}, publisher = {Association for Computational Linguistics}, url = {https://aclanthology.org/2025.findings-acl.1033/}, doi = {10.18653/v1/2025.findings-acl.1033}, keywords = {ai4health}, } -
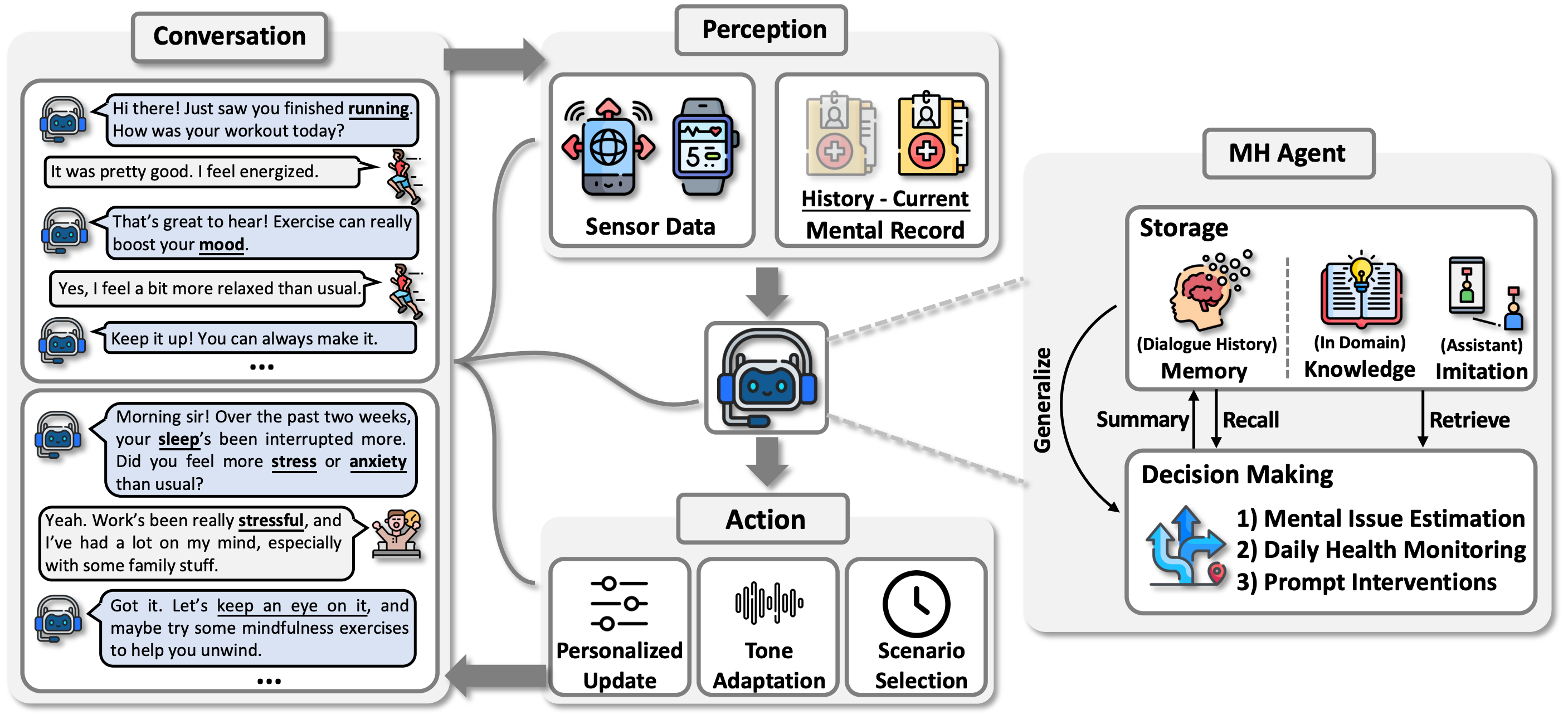 Transforming mental health care with autonomous llm agents at the edgeSijie Ji*, Xinzhe Zheng*, Wei Gao, and 1 more authorIn ACM Sensys, 2025
Transforming mental health care with autonomous llm agents at the edgeSijie Ji*, Xinzhe Zheng*, Wei Gao, and 1 more authorIn ACM Sensys, 2025The integration of Large Language Models (LLMs) with mobile devices is set to transform mental health care accessibility and quality. This paper introduces MindGuard, an autonomous LLM agent that utilizes mobile sensor data and engages in proactive, personalized conversations while ensuring user privacy through local processing. Unlike traditional mental health AI tools, MindGuard enables real-time, context-aware interventions by dynamically adapting to users’ emotional and physiological states. The real-world implementation demonstrates its effectiveness with the ultimate goal of creating an accessible, scalable, and personalized mental healthcare ecosystem for anyone with smart mobile devices.
@inproceedings{ji2025transforming, keywords = {ai4health}, title = {Transforming mental health care with autonomous llm agents at the edge}, author = {Ji, Sijie and Zheng, Xinzhe and Gao, Wei and Srivastava, Mani}, booktitle = {ACM Sensys}, pages = {692--693}, year = {2025}, }
2024
-
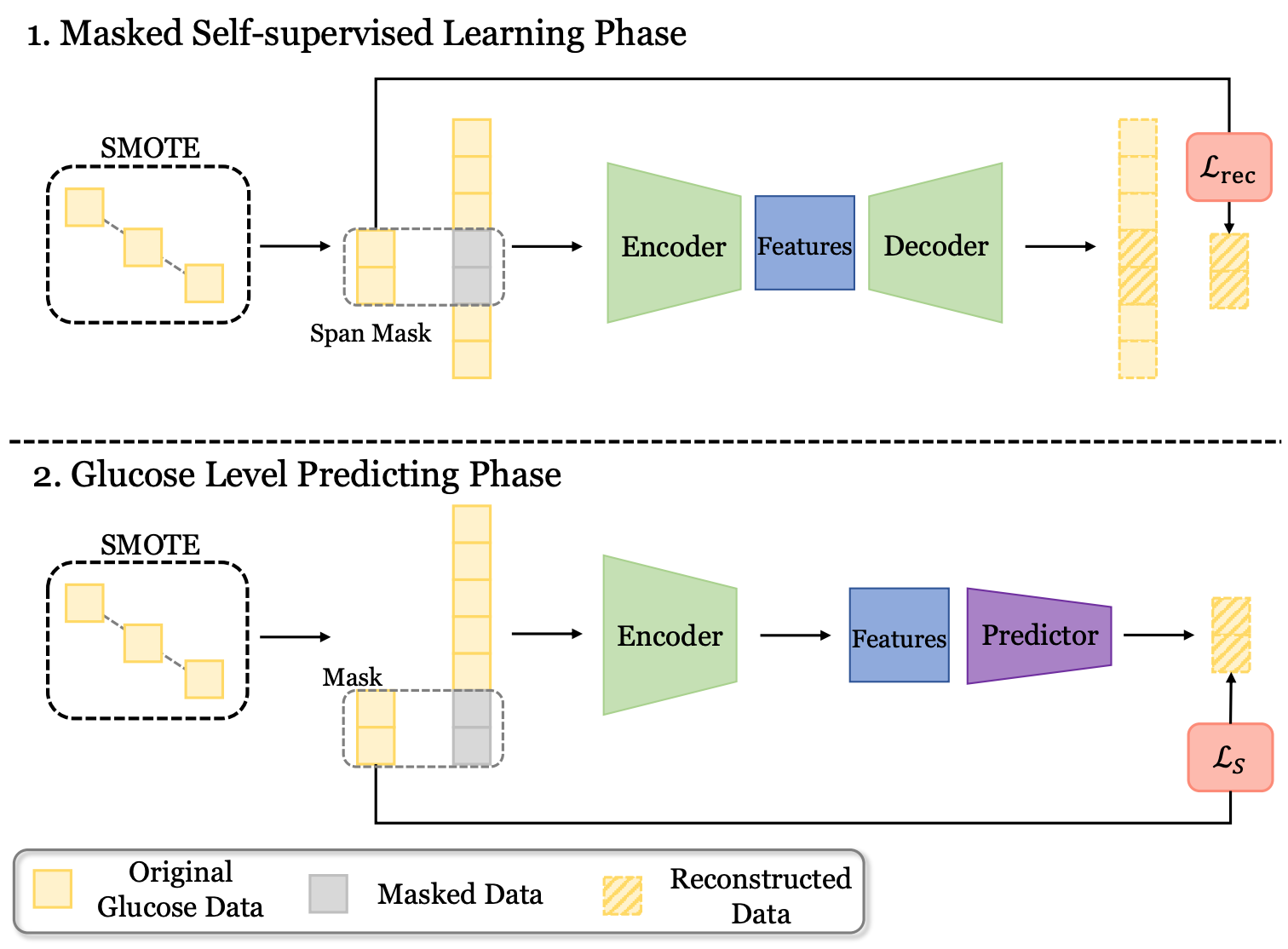 Predicting adverse events for patients with type-1 diabetes via self-supervised learningXinzhe Zheng, Sijie Ji, and Chenshu Wu†In ICASSP, 2024
Predicting adverse events for patients with type-1 diabetes via self-supervised learningXinzhe Zheng, Sijie Ji, and Chenshu Wu†In ICASSP, 2024Predicting blood glucose levels is fundamental for precise primary care of type-1 diabetes (T1D) patients. However, it is challenging to predict glucose levels accurately, not to mention the early alarm of adverse events (hyperglycemia and hypoglycemia), namely the minority class. In this paper, we propose BG-BERT, a novel self-supervised learning framework for blood glucose level prediction. In particular, BG-BERT incorporates masked autoencoder to capture rich contextual information of blood glucose records for accurate prediction. More specifically, SMOTE data augmentation and shrinkage loss are employed to effectively handle adverse events without discrimination. We evaluate BG-BERT on two benchmark datasets against two state-of-the-art base-line models. The experimental results highlight the significant improvements achieved by BG-BERT in glucose level prediction accuracy (measured by RMSE) and sensitivity to adverse events, with average lifting ratios of 9.5% and 44.9%, respectively.
@inproceedings{zheng2024predicting, title = {Predicting adverse events for patients with type-1 diabetes via self-supervised learning}, author = {Zheng, Xinzhe and Ji, Sijie and Wu, Chenshu}, booktitle = {ICASSP}, pages = {1526--1530}, year = {2024}, organization = {IEEE}, keywords = {ai4health}, }
Mobile Computing
I no longer work in this area, but here are some of my past publications.
2025
-
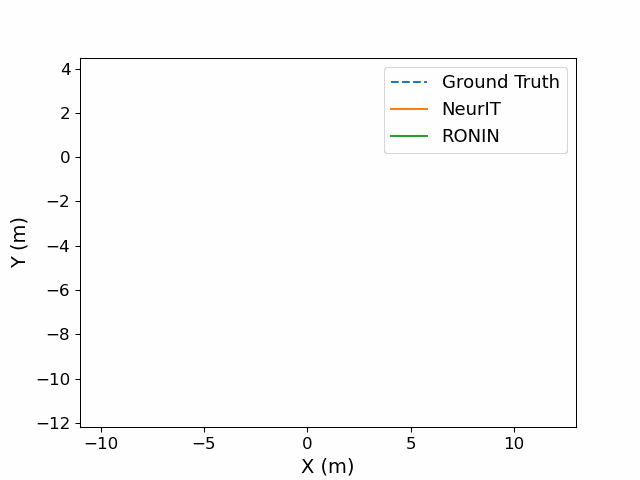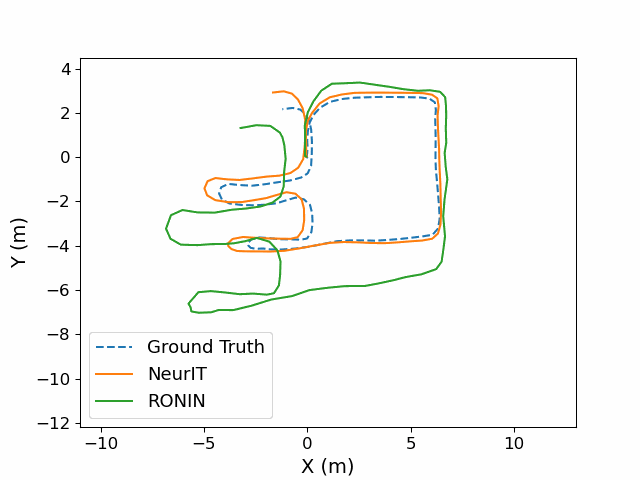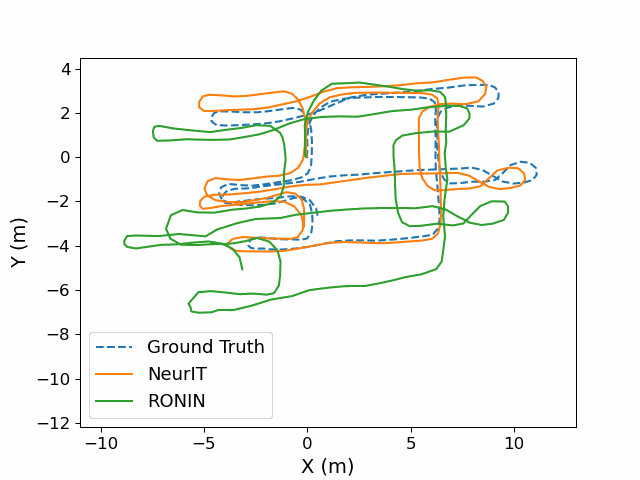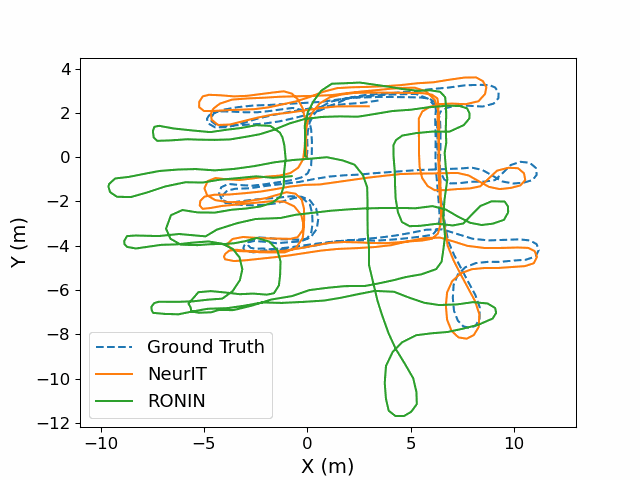 NeurIT: Pushing the Limit of Neural Inertial Tracking for Indoor Robotic IoTXinzhe Zheng, Sijie Ji, Yipeng Pan, and 2 more authorsIEEE Transactions on Mobile Computing, 2025
NeurIT: Pushing the Limit of Neural Inertial Tracking for Indoor Robotic IoTXinzhe Zheng, Sijie Ji, Yipeng Pan, and 2 more authorsIEEE Transactions on Mobile Computing, 2025Inertial tracking is vital for robotic IoT and has gained popularity thanks to the ubiquity of low-cost inertial measurement units and deep learning-powered tracking algorithms. Existing works, however, have not fully utilized IMU measurements, particularly magnetometers, nor have they maximized the potential of deep learning to achieve the desired accuracy. To address these limitations, we introduce NeurIT, which elevates tracking accuracy to a new level. NeurIT employs a Time-Frequency Block-recurrent Transformer (TF-BRT) at its core, combining both RNN and Transformer to learn representative features in both time and frequency domains. To fully utilize IMU information, we strategically employ body-frame differentiation of magnetometers, considerably reducing the tracking error. We implement NeurIT on a customized robotic platform and conduct evaluation in various indoor environments. Experimental results demonstrate that NeurIT achieves a mere 1-meter tracking error over a 300-meter distance. Notably, it significantly outperforms state-of-the-art baselines by 48.21% on unseen data. Moreover, NeurIT demonstrates robustness in large urban complexes and performs comparably to the visual-inertial approach (Tango Phone) in vision-favored conditions while surpassing it in feature-sparse settings. We believe NeurIT takes an important step forward toward practical neural inertial tracking for ubiquitous and scalable tracking of robotic things. NeurIT is open-sourced here: https://github.com/aiot-lab/NeurIT.
@article{zheng2025neurit, title = {NeurIT: Pushing the Limit of Neural Inertial Tracking for Indoor Robotic IoT}, author = {Zheng, Xinzhe and Ji, Sijie and Pan, Yipeng and Zhang, Kaiwen and Wu, Chenshu}, journal = {IEEE Transactions on Mobile Computing}, year = {2025}, publisher = {IEEE}, keywords = {mobilecomputing}, } -
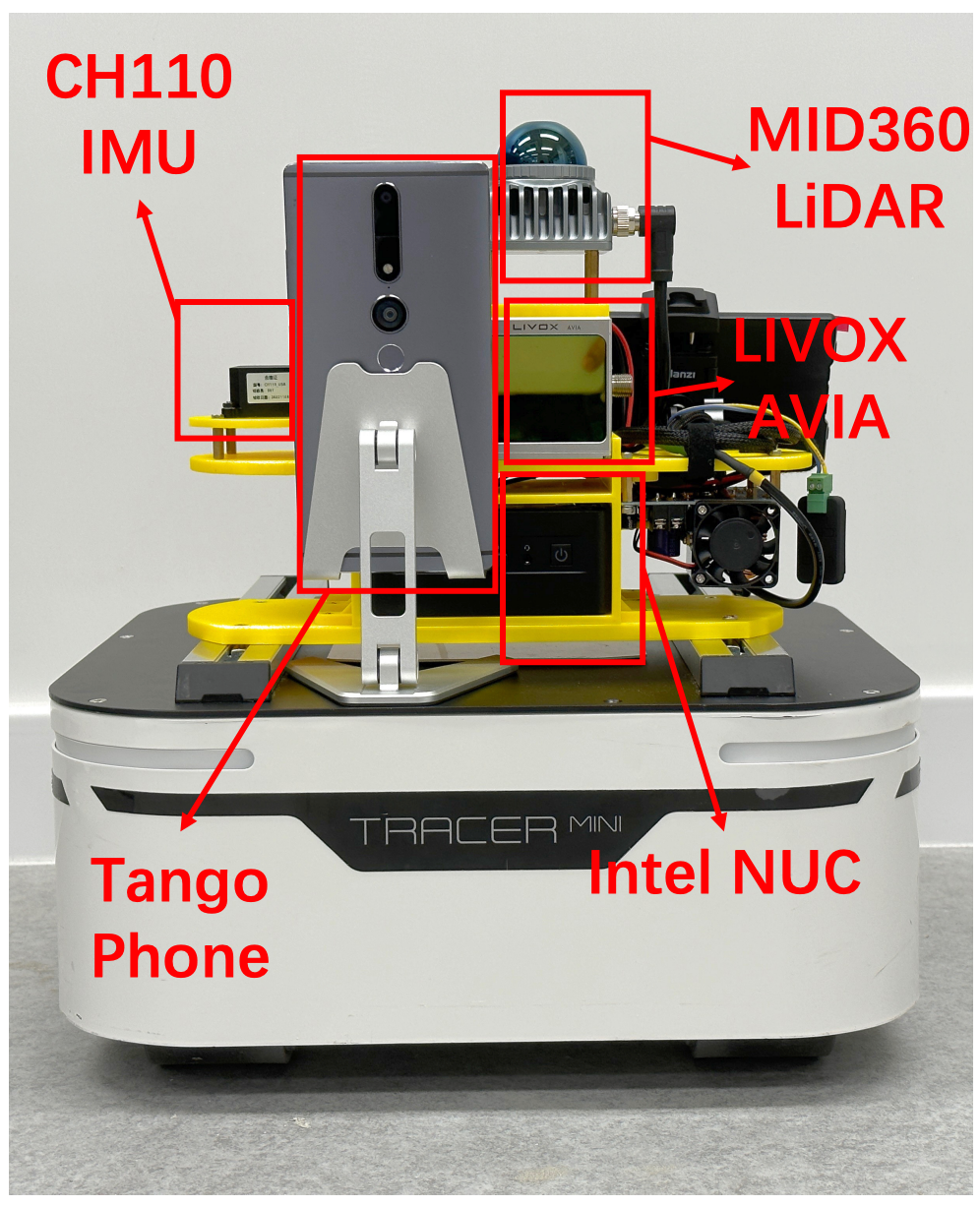 Magnetometer-Calibrated Hybrid Transformer for Robust Inertial Tracking in RoboticsXinzhe Zheng, Sijie Ji, Yipeng Pan, and 3 more authorsIn ICRA, 2025
Magnetometer-Calibrated Hybrid Transformer for Robust Inertial Tracking in RoboticsXinzhe Zheng, Sijie Ji, Yipeng Pan, and 3 more authorsIn ICRA, 2025Inertial tracking is vital for autonomous robots and has gained popularity with the ubiquity of low-cost Inertial Measurement Units (IMUs) and deep learning-powered tracking algorithms. Existing works, however, have not fully utilized IMU measurements, particularly magnetometers, nor maximized the potential of deep learning to achieve the desired accuracy. To bridge the gap, we introduce NeurIT, which employs a Time-Frequency Block-recurrent Transformer (TF-BRT) at its core, combining RNN and Transformer to learn both time-frequency representative features. To fully utilize IMU information, we strategically employ differentiation of body-frame magnetometers for orientation calibration in a sensor fusion manner. Experiments conducted in diverse environments show that NeurIT maintains a mere 1 -meter tracking error over a 300 - meter distance, surpassing state-of-the-art baselines by 48.21 % on unseen data. NeurIT also performs comparably to the visual-inertial approach (Tango Phone) in vision-favored conditions and surpasses it in plain environments. We share the code and data to promote further research: https://github.com/aiot-lab/NeurIT.
@inproceedings{zheng2025magnetometer, title = {Magnetometer-Calibrated Hybrid Transformer for Robust Inertial Tracking in Robotics}, author = {Zheng, Xinzhe and Ji, Sijie and Pan, Yipeng and Zhang, Kaiwen and Pan, Jia and Wu, Chenshu}, booktitle = {ICRA}, pages = {16102--16108}, year = {2025}, organization = {IEEE}, keywords = {mobilecomputing}, }
2024
-
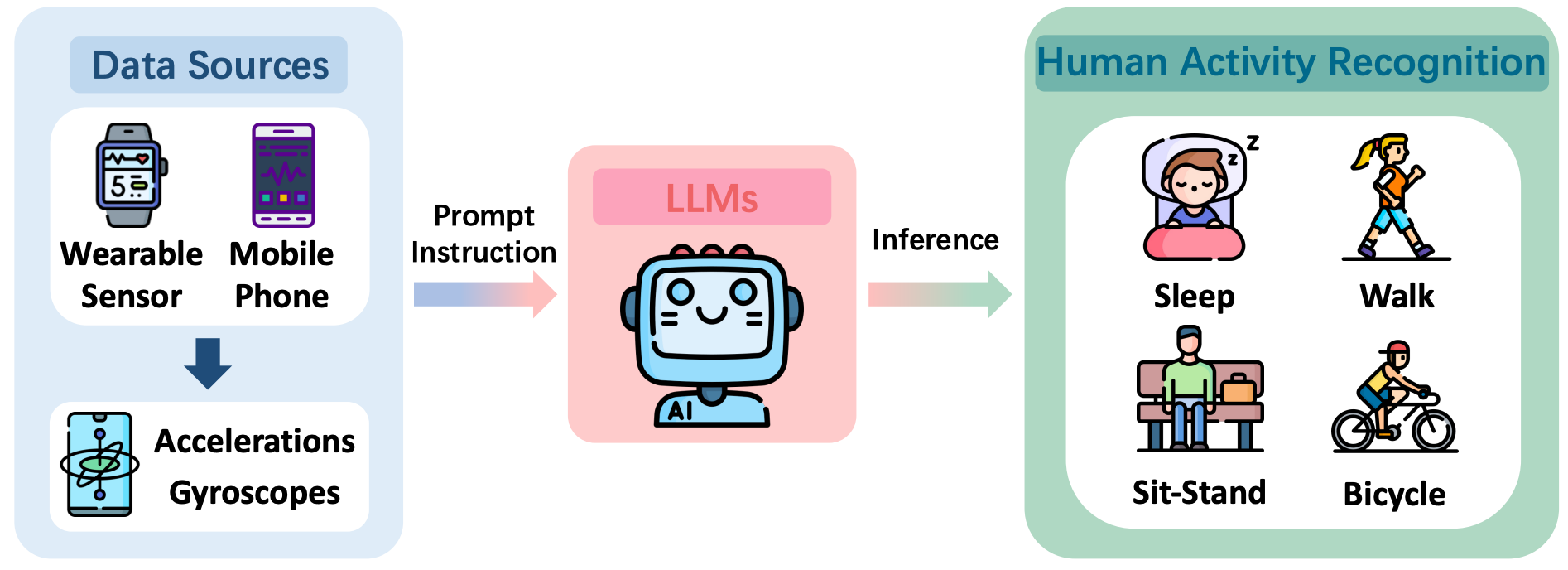 Hargpt: Are llms zero-shot human activity recognizers?Sijie Ji*, Xinzhe Zheng*, and Chenshu Wu†In IEEE FMSys, 2024
Hargpt: Are llms zero-shot human activity recognizers?Sijie Ji*, Xinzhe Zheng*, and Chenshu Wu†In IEEE FMSys, 2024There is an ongoing debate regarding the potential of Large Language Models (LLMs) as foundational models seamlessly integrated with Cyber- Physical Systems (CPS) for interpreting the physical world. In this paper, we carry out a case study to answer the following question: Are LLMs capable of zero-shot human activity recognition (HAR)? Our study, HARGPT, presents an affirmative answer by demonstrating that LLMs can comprehend raw IMU data and perform HAR tasks in a zero-shot manner, with only appropriate prompts. HARGPT inputs raw IMU data into LLMs and utilizes the role-play and “think step-by-step“ strategies for prompting. We benchmark HARGPT on GPT4 using two public datasets of different inter-class similarities and compare various baselines both based on traditional machine learning and state-of-the-art deep classification models. Remarkably, LLMs successfully recognize human activities from raw IMU data and consistently outperform all the baselines on both datasets. Our findings indicate that by effective prompting, LLMs can interpret raw IMU data based on their knowledge base, possessing a promising potential to analyze raw sensor data of the physical world effectively.
@inproceedings{ji2024hargpt, title = {Hargpt: Are llms zero-shot human activity recognizers?}, author = {Ji, Sijie and Zheng, Xinzhe and Wu, Chenshu}, booktitle = {IEEE FMSys}, pages = {38--43}, year = {2024}, organization = {IEEE}, keywords = {mobilecomputing}, }
2023
- ScamRadar: Identifying Blockchain Scams When They are PromotingXinzhe Zheng, Pengcheng Xia, Kailong Wang, and 1 more authorIn International Conference on Blockchain and Trustworthy Systems, 2023
2022
- Resource allocation on blockchain enabled mobile edge computing systemXinzhe Zheng, Yijie Zhang, Fan Yang, and 1 more authorElectronics, 2022